|
Size: 2002
Comment:
|
Size: 1843
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 20: | Line 20: |
| White stls - segmentation from MRI. Colored bone stls - progression throughout test (red = initial, green = 1/4 way through test, blue = 1/2 way through test). Colored registration marker stls - spheres from calculated center of digitized points. <<BR>> {{attachment:OK_reg1.png||height=300}} {{attachment:OK_reg2.png||height=300}} |
White stls - segmentation from MRI. Colored registration marker stls - spheres from calculated center of digitized points. <<BR>> {{attachment:OK_reg1.png||height=300}} |
| Line 29: | Line 29: |
| <<EmbedObject(005_passive flexion 0-60_main_processed.mpeg, width=637px,height=392px,play=true,loop=true)>> | See [[attachment:005_Passive Flexion 0-60_main_processed.mp4|Sample Passive Flexion Video]] for video example. |
Target
- Create visualization of experimental tests in modeling (image) coordinate system.
- Prepare experimental data for model inputs.
Registration
- Download joint mechanics .zip file from Downloads page.
- Download the desired registration markers and bone STLs (place in folder named mri-oks00x, where x is the specimen ID).
- 3 registration marker stls for tibia
- 3 registration marker stls for femur
- stl for tibia bone
- stl for femur bone
Create registration xml (Example Registration XML)
Run script from command line Registration Script
Change directory to desired OpenKnee specimen (e.g. oks001)
Run convert_to_image_CS.py <oks00x_registration_0x.xml> <1>
Example using oks001 - 004_passive flexion
White stls - segmentation from MRI. Colored registration marker stls - spheres from calculated center of digitized points.
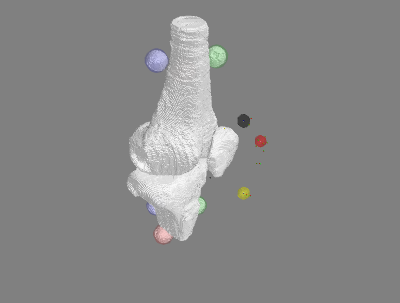
Re-sample Data for Model Input
- Create text file with desired tdms file(s) you would like to re-sample
Place in the appropriate directory KinematicsKinetics of either the TibioFemoral or PatelloFemoral test directories.
Run script from command line Re-sampling Script
Change directory to desired OpenKnee specimen (e.g. oks001)
Run resampling_potting.py <oks00x_registration_0x.xml> <oks0x_xF.txt>
See Sample Passive Flexion Video for video example.