Contents
Target Outcome
Prerequisites
Protocols
File Association
- Setting up the Directory
- Download and unpack the subject folder and place in MULTIS_trials folder
- The folder/file configuration should be the following
MULTIS_trials
MULTIS###-# (Subject folder containing all data)
Configuration - Configuration files and subject XML
Data - All "tdms" files
Ultrasound - All "dicom" files
PulseWidths300.csv - contains key to decode file association pulse
manualMatcherFA.py
plotterFA.py
XMLparser.py
dataAnalysis.py
tdsmParserMultis.py
- Running the File Association script
- Install libraries as needed
- Change Directory to match desired Subject ID
- Ensure Plot is True to save all verification plots
- Run script.
- Check directory in file browser under the subject folder for the following additions
- FileAssociationPNG - folder
TimeSynchronization.txt - file containing trial numbers and delta T's of accepted trials only
- MULTIS###-#readme.txt - human readable file which summarizes the file matching
- Manually Matching
- Open the 'MULTIS###-#readme.txt' file
- Note the Unmatched and Accepted Trials, specifically the binary column
- Examine any files beginning with 'NMa', meaning no match found
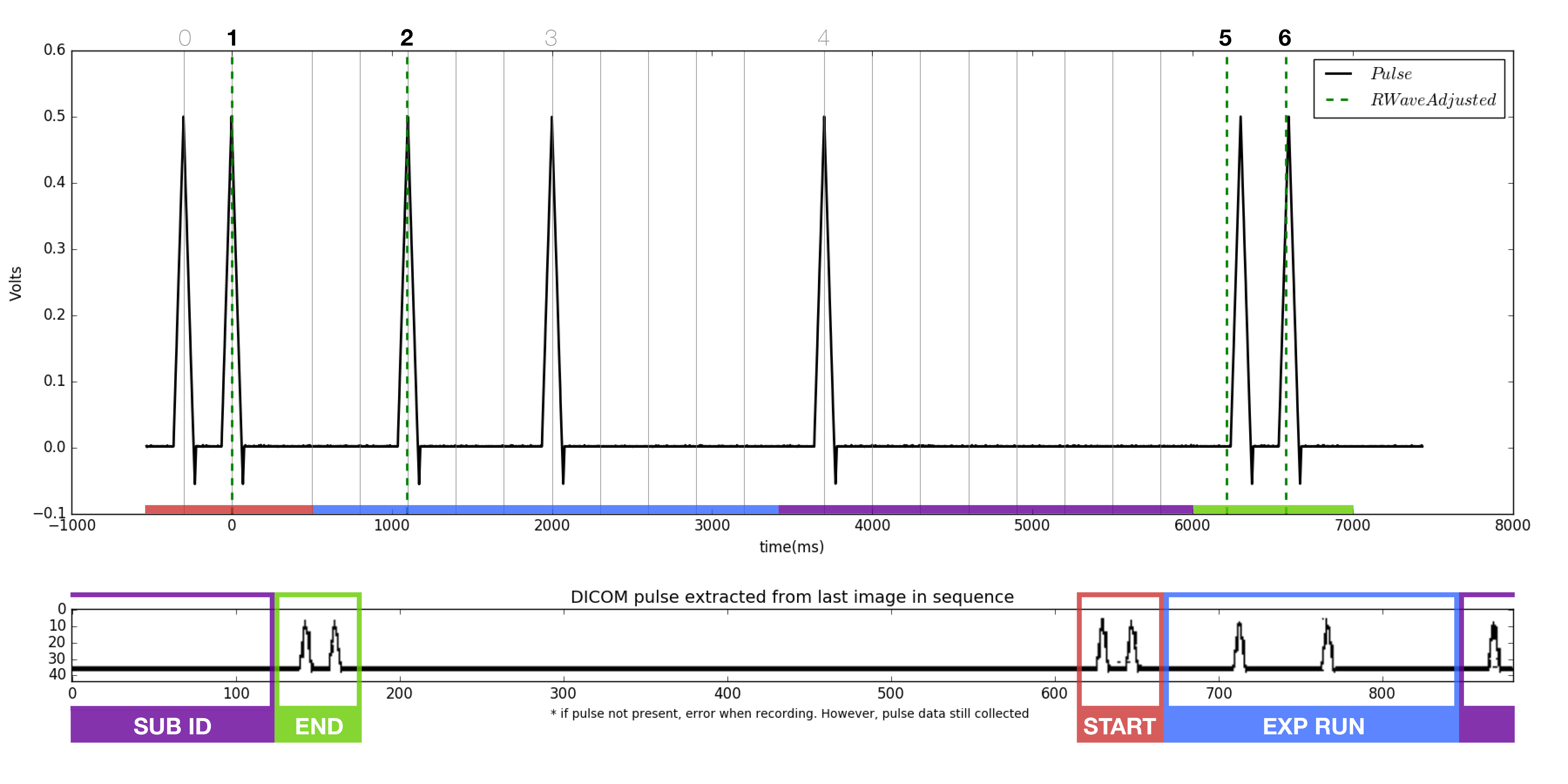
- Locate the start and end pluses, each a burst of two peaks.
- Compare the binary of the trial run number to each of the pulse sequences found in the 'NMa' png's. Note the file names of each. At this point, it is known which file each of the missing accepted trial numbers belong to.
Open the manualMatcherFA.py and plotterFA.py python scripts.
Change the directory file names in the manualMatcherFA script to a pair of matching files found in step 2. Only one pair of files will be compared at a time. The files will be imported to the plotterFA script automatically.
- Run the plotterFA.py script, ensuring iteration is set to zero.
- Examine the figure generated.
- The pulse extracted from the dicom (bottom of figure) should match the pulse from the tdms file (top of figure). Note the dicom pulse will not always begin at the left most portion of the window as it is a continuous loop in the ultrasound machine.
- As before, locate the beginning and ending double pulses.
- Count the number of peaks present in the dicom pulse.
- Count the number of RWaves in the generated figure.
- It is likely that one or more of the RWaves are missing, as that is why the files did not match.
- Change the iteration from 0, to 1, to 2, and so on and re-plot each time until the RWaves appear to line up with the pulse peaks in the top figure.
- Identify which peaks have RWaves, and which do not.
- Each of the peaks in the upper figure has a number associated with it in the script. Starting from 0, instead of 1, the peaks are named accordingly.
- Record which peaks have RWaves. For example. (1,2,5,6) may have RWaves.
- Open the 'MULTIS###-#readme.txt' file
Return to manualMatcherFA.py
- Ensure iteration is set to 2.
- Change the chosenRwaves variable to reflect the the recorded RWaves i.e. (1,2,5,6)
- Run the script
- Check the following items
- The filename of the dicom and tdms changed appropriately
- The trial number and time synch were added to
MULTIS###-#readme.txt
The updated PNG was added to the FileAssociationPNG folder
Quality Assurance
- Setting up the Directory
- same as file association with the addition of folders created by file association
- Running the dataAnalysis.py script
- Change Directory to reflect subject ID
- Run script
- Check the following
A folder named AnalysisPNG should appear under MULTIS###-#
- 56 png files should be found within this folder
- Analysis
- There are two types of figures
- Anatomical
- 3 dicom images (at 1st pulse, minimum force, last pulse) over load cell data
- Indentation
- 4 dicom images (at pre, start, middle, max indentation) over load cell data
- Anatomical
- There are two types of figures

