Contents
Naming Convention
- 019_SMULTIS001-1_SXX_CUT_SKN
- TRI_SUBJECTID_TOOL_ACTION_LOC
TOOL
- SXX-Scalpel
- FXX-Forceps
- RXX-Retractor
- SFX-Scalpel/Forceps
ACTION
- CUT = CUT
- EVE = EVERT
- WID = WIDEN
- CLO = CLOSURE
- RET = RETRACT
- SUT = SUTURE
- DIS = DISSECT
- PCH = PINCH
LOCATION
- SKN = SKIN
- SFI = SKIN FAT INTERFACE
- FAT = FAT
- FMI = FAT MUSCLE INTERFACE
- MUS = MUSCLE
- MBI = MUSCLE BONE INTERFACE
Supplies
- 3 instrumented tools
- Scalpel + Indenter
- Forceps
- retractor
- 2 prong retractor for skin
- full size retractor for fat and muscle
- 2 allen wrenches to change scalpel blades
- 5 scalpel blades #15
- 2 4-0 sutures
- sharpie
- ruler
- calipers
Protocols
Strain Measurement
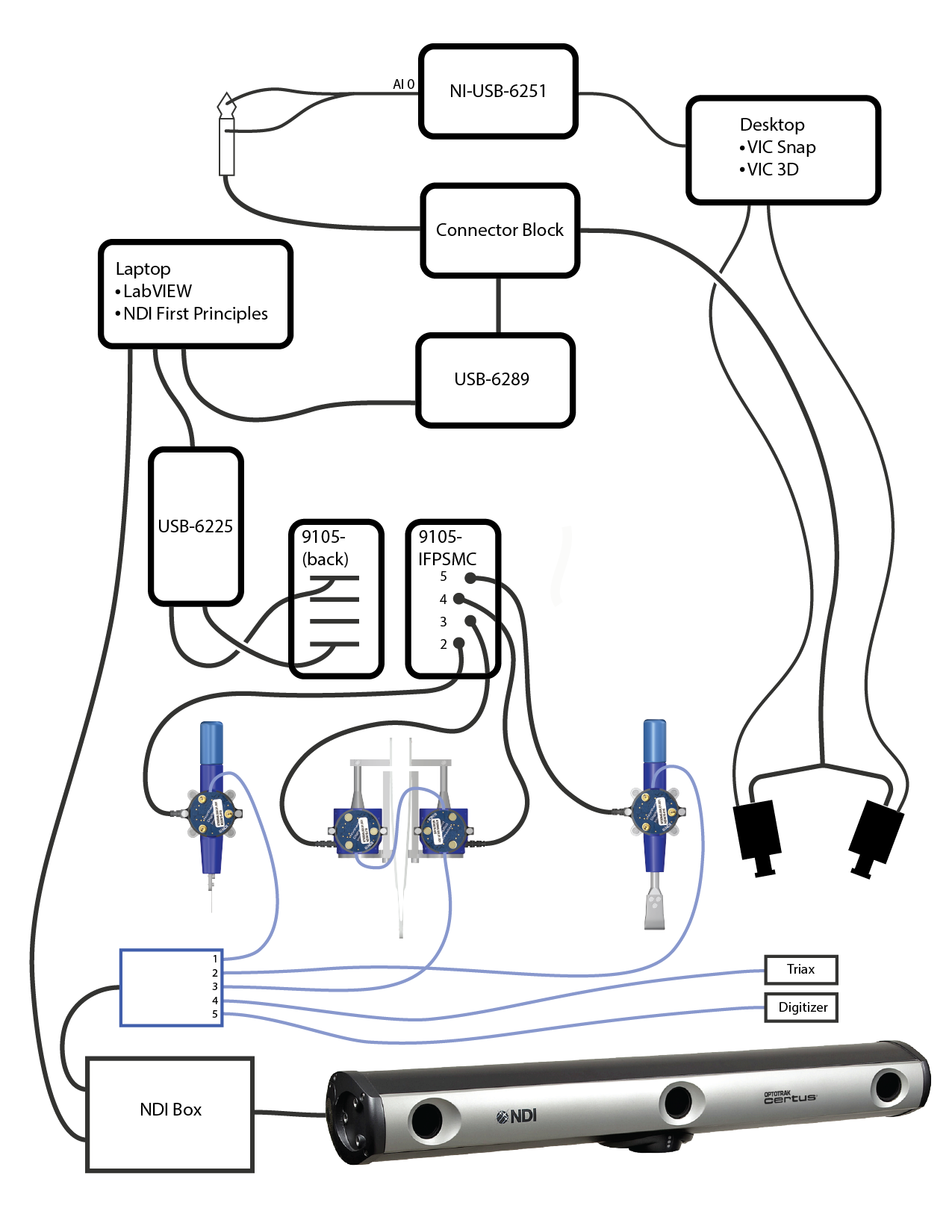
Hardware
- Connect NI-USB-6289 to multis laptop.
- Connect SCB 68A terminal hub to NI-USB-6289
- Connect camera trigger to SCB 68A
- Connect both yellow wires to pins 55 and 66
- Connect both green wires to pins 33 and 22
- Both green and yellow wires are attached to a BCB cable
- The BCB cable connects to a splitter, which connects to each Vic3D camera
- Connect analog synchronization signal to SCB 68A using instrumented ultrasound setup
- Use Connector 1 to connect to SCB 68A
- Connect the 1/4 stereo jack to a BCB cable
- Connect BCB cable to AI0 of the NI USB 6251
- Connect the NI USB 6251 to the Vic3D computer
Software
- Open Vic-Snap
- Set appropriate project path and Speckle name, the calibration name will automatically update
- Click 'calibration images' button
Click Images > Synch Mode > Hardware
Click Images > TTL Capture > Low (trigger level) > Streaming (Capture)
- Click Arm when ready (in TTL Capture window)
- When armed and in Speckle images mode, the Multis computer will trigger capture and send synchronization signal simultaneously
Camera Setup
- Set focus manually to surface of skin
- Right click in camera viewport and select 'show focus/contrast'
- Connect iPad (connected to wifi dongle MultisVIC3d network)
- Show cross hairs on iPad and pick a point to align the center of each crosshair. This should be the center of the show focus contrast region.
- Adjust polarized lens on each camera and light
- First optimize polarized lenses for undeformed state (maximum purple on focus/contrast)
- Next rotate each filter on the light sources 90 degrees so deformed tissue will not cause glare
- Mark corners of specimen fixture with tape,
- Push specimen fixture forward to make room for calibration
- Calibrate by holding appropriately sized calibration plate at level of focus (leg surface) and capturing images while pivoting in all directions (20 images)
- Select biggest calibration plate that will fit in camera view (cannot exceed)
Speckle Application
- Shave and clean surface of skin
- Apply foundation to mute surface reflection
- Prepare the rubber speckle pad with ink, apply 3 times in 3 different orientations
Surgical Procedures
The following procedures are to be tested using the instrumented surgical tools. After each cut, tissue will be retracted for following cut.
- TARE load cells
- Place 3 dots at 0, 5, and 10cm in the target region after speckling
- 0-Indent Skin - w/ Strain measurement
- SXX_IND_SKN
- Location: 5cm dot
- Approach from lateral side of leg
- 1-Pinch Skin - w/ Strain measurement
- FXX_PCH_SKN
- Location: 5cm dot
- Approach from lateral side of leg. Start pinch with 3 mm width, pull gently upwards to 3-5mm
- TARE load cells
- 2-Cut Skin - w/ Strain measurement
- SXX_CUT_SKN
- Tool: Scalpel #15 blade
- Location: 0 to 10cm dots
- Procedure: Cut 10cm straight line in longitudinal bone direction (proximal to distal) at anterior central location of leg
- Blade Orientation: Face perpendicular to tissue, cutting edge 45deg to tissue
- Approach incision start by moving distal to proximal, cut proximal to distal (for all cutting). Place left hand to on proximal side of incision to stabilize the tissue.
- Measure depth, length, width of cut
- Approach SFI but do not puncture (non-instrumented)
- Measure depth, length, width of cut
- Attach new scalpel blade
- 3-Cut Skin/Fat interface
- SXX_CUT_SFI
- Tool: Scalpel #15 blade
- Procedure: Cut 10cm straight line in direction of first incision in same orientation. Place left hand to on proximal side of incision to stabilize the tissue.
- Measure depth, length, width of cut
Note: Orient forceps in each trial with left tip near center of incision
- 4-Forceps - Widening
- FXX_WID_SFI
- Tool: Forceps
- Procedure: Pinch tissue at incision, widen tissue 2 cm (central to medial)
- Purpose: imitate opening of incision to explore region
- FXX_WID_SFI
- Measure depth, length, width of cut
- 5-Forceps - Closing approximation
- FXX_CLO_SFI
- Tool: Forceps
- Procedure: Pinch tissue at incision, bring incision together (close it)
- Purpose: pulling tissue together after incision to approximate edges before suturing
- 6-Forceps - Everting
- FXX_EVE_SFI
- Tool: Forceps
- Procedure: Pinch tissue at incision, lift tissue will twisting forceps (clockwise) and opening incision apprx 1-2 cm
- Purpose: evert tissue for suture insertion
- Measure depth, length, width of cut
- TARE load cells
- 7-Suturing: Skin layer w/ SFI
- SXX_SUT_SFI
- Note: Prepare suture up to point of closure of first knot, but do not close
- attach one end to instrumented scalpel (remove blade and tie around bolt using a surgeon's knot)
- grasp non-instrumented forceps with the thread from the instrumented end
- pull both ends, one with non-instrumented, the other with instrumented, to complete first knot.
- Pulling Direction: orient instrumented attachment to lateral side (pull laterally)
- Cut and remove suture
- Measure depth, length, width of cut
- TARE load cells
- 8-Cut Fat
- SXX_CUT_FAT
- Tool: Scalpel
- Procedure: Cut fat only to a depth 3/4 the length of the blade, 10cm distance
- 9-Retract Fat
- RXX_RET_FAT
- Tool: Retractor
- Procedure: Insert retractor, pull skin and fat away from incision 2cm. Use non-instrumented retractor for opposing side
- Pulling Direction: Parallel to surface and perpendicular to incision, medial to lateral
- Measure depth, length, width
- Approach FMI (non-instrumented)
- Attach new scalpel blade
- Measure depth, length, width
- 10-Cut through FMI into Muscle
- SXX_CUT_FMI
- Tool: Scalpel #15 blade
- Procedure: While retracting, cut 10cm straight line in direction of first incision in same orientation to a depth 3/4 the length of the blade
- Measure depth, length, width
- 11-Retract Muscle
- RXX_RET_MUS
- Tool: Retractor
- Procedure: use 9-Retract Fat
- Measure depth, length, width
- Attach new scalpel blade
- 12-Cut Muscle
- SXX_CUT_MUS
- Tool: Scalpel #15 blade
- Procedure: Cut muscle only to a depth half the length of the blade, 10cm distance
- Measure depth, length, width
- 13-Retract Muscle
- RXX_RET_MUS
- Tool: Retractor
- Procedure: use 9-Retract Fat
- Measure depth, length, width
- Cut to bone (non-instrumented)
- Measure depth, length, width
- 14-Retract Muscle (include all tissue down to bone surface)
- RXX_RET_MBI
- Tool: Retractor
- Procedure: use 9-Retract Fat
- Measure depth, length, width
- 15-Forceps Dissection Fat and Muscle
- SFX_DIS_FMI
- Tool: Forceps + Scalpel
- Procedure: Pinch tissue at midpoint of incision (medial side of incision), open incision and separate tissue with scalpel
- Measure depth, length, width
- 16-Forceps Dissection Skin and Fat
- SFX_DIS_SFI
- Tool: Forceps + Scalpel
- Procedure: Pinch tissue at midpoint of incision (medial side of incision), open incision and separate tissue with scalpel using the following protocols
- Measure depth, length, width
Data transfer
- On the load cell computer, zip the raw data for this subject. The folder can be found in c:\sMULTIS data.
The zipped folder can be transferred to MIDAS (http://cobicore.lerner.ccf.org/midas/community/7) using the secure wireless connection. The data will be saved in the path /Private. The folder may be unzipped, but leave original zipped folder.
- VIC3D data is first compressed on the VIC3D computer under the Documents folder. It must be transferred to an external hard drive and uploaded via a networked computer to MIDAS under the relevant donorID folder, where it remains zipped.
Output
All raw data can be found at http://cobicore.lerner.ccf.org/midas/community/7
- Configuration and data directories are located in the path /Private/SMULTISXXX-1/, where XXX is the unique subject identifier (i.e., 001, 002, 003, etc.)
Next step: DataAnalysis