Target Outcome
The goal of this data analysis specification is to provide tissue thicknesses of skin, fat and muscle for both in vivo and in vitro experimentation. The output data can then be used for modeling purposes, statistical data analysis, and comparison to MRI and CT thicknesses.
Prerequisites
Infrastructure
Previous Protocols
Requirements
- Python [Version 2.7]
Procedures
Ultrasound Thickness Analysis
Thickness Analysis Software
Version 1.0 places red circles at the coordinates of the previous frame that was analyzed, starting with the second frame. Version 1.1 moves red circles to the location with the closest matched distribution to the coordinates in the previous frame, starting with the second frame. Version 2.0 does not require the user to hit 'enter' to save the thicknesses and allows the user to cycle through all of the locations for one subject instead of restarting the program after each location.
- Setting up the Directory
- Download and unpack the desired Subject ID folder, make sure you use the 'associated' data if you do not want to associate the files yourself.
Note: If you have just completed the file association, confirm that the dicom and tdms files within the Ultrasound and Data folders have been matched (should begin with a three digit number) and that the corresponding deltaT values (as xml files) are located in the TimeSynchronization folder.
- Download and unpack the desired Subject ID folder, make sure you use the 'associated' data if you do not want to associate the files yourself.
- The folder/file configuration should be the following
MULTIS###-# (Subject folder containing all data - the required data for the thickness measurements is shown below)
Configuration - Contains subject xml file for accepted trials
Data - Contains tdms files
Ultrasound - Contains dicom files
TimeSynchronization - Contains xml files with deltaT values for matched trials
- Required Python scripts
TA_sfm_v#.#.py
tdsmParserMultis.py
XMLparser.py
Plot_thicknessForce.py
- Running the manual thickness measurement script
- Install libraries as needed
- Change the directory to match where the Python scripts are located
- Run script
- The first dialog window will allow you to select the Subject folder (you may also choose to hard code the path to the Subject folder, found within the getFiles() function)
- Browse through to find the location of this folder and click 'ok'
- A list will appear in another window which will show all the trials for that subject (you may have to scroll down to see the entire list)
- Double-click on the location you would like to analyze
- After a couple of seconds, the first image will appear
- The program behaves differently depending on if the trial is anatomical or indentation
- Anatomical - The program begins with the frame corresponding to the lowest resultant force
- Indentation - The program begins with the frame that corresponds to the beginning of indentation
- The program behaves differently depending on if the trial is anatomical or indentation
- Drag the red dots so that the center of the dot is over each boundary (use the built in matplotlib toolbar to zoom in if needed)
- Transducer/Skin boundary
- Skin/Fat boundary
- Fat/Muscle boundary
- Muscle/Bone boundary
- When you are happy with your selection, press 'next image' and the results will be calculated and saved to an xml file.
- The next frame for analysis will automatically appear
- Anatomical trials will give you the image corresponding to the next lowest force, until all frames within the first pulse and last pulse range have been analyzed
- Indentation trials will give you the next image in the sequence until reaching the maximum force
- After analyzing the frames for that location, hit the 'Done' button and the list of images for that subject will re-appear and you can analyze a new location.
- The xml file that was saved will be re-formatted using pretty print (only if you hit the 'Done' button)
A PNG folder within the Analysis folder will be created that has saved images of the first frame that was analyzed as well as a graph showing the force/thickness relationships
- You may end the program at any time by clicking the 'X' button in the upper right hand corner.
- All windows will be closed and the script will stop running
- All windows will be closed and the script will stop running
- The first dialog window will allow you to select the Subject folder (you may also choose to hard code the path to the Subject folder, found within the getFiles() function)
- Data storage
- The xml file will be named using the tdms filename with '_manThick'+ time stamp appended
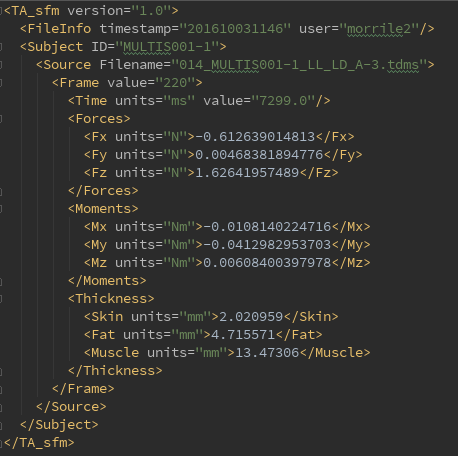
- Version of software used for analysis (TA_sfm.py)
- File Info includes the time stamp at the beginning of analysis as well as the user name at the time of program execution
- Subject shows the MULTIS ID name
- Source shows the Filename of the data (tdms) file
- A new 'Frame' child is appended for each frame that is analyzed
- The Frame child includes the following information:
- Value - Frame number
- Time - Reference time from the ultrasound time vector data (in ms)
- Forces - Forces in the x, y, and z directions (in N)
- Moments - Moments in the x, y, and z directions (in Nm)
- Thicknesses - Skin, Fat, and Muscle layers (in mm)
- The Frame child includes the following information:
- The xml file will be named using the tdms filename with '_manThick'+ time stamp appended
Thickness Inclusion XML
- A single XML document was also developed to streamline the process of searching for the appropriate thickness XMLs. This file is used for data analysis scripts as well as modeling scripts.
Repeatability & Reproducibility
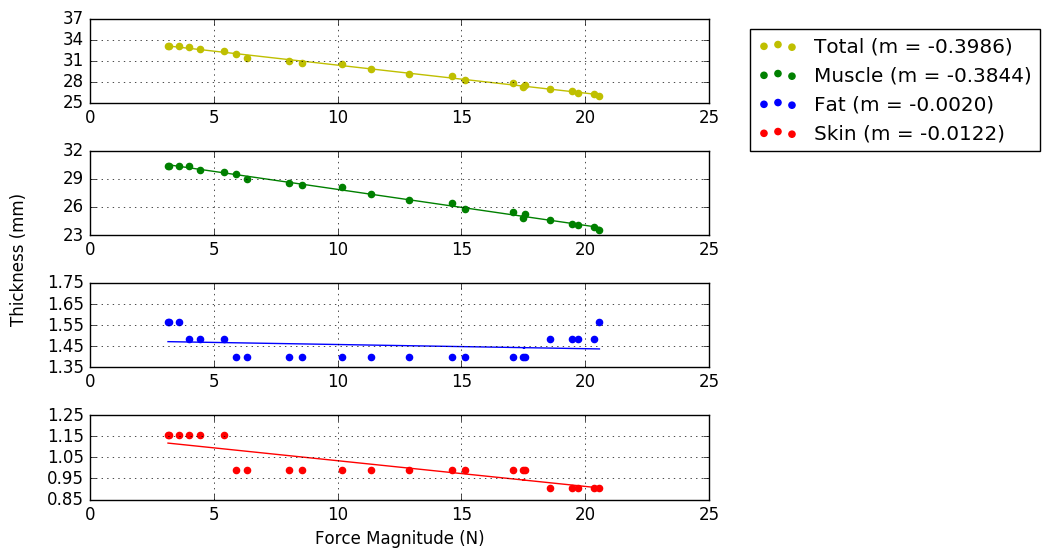
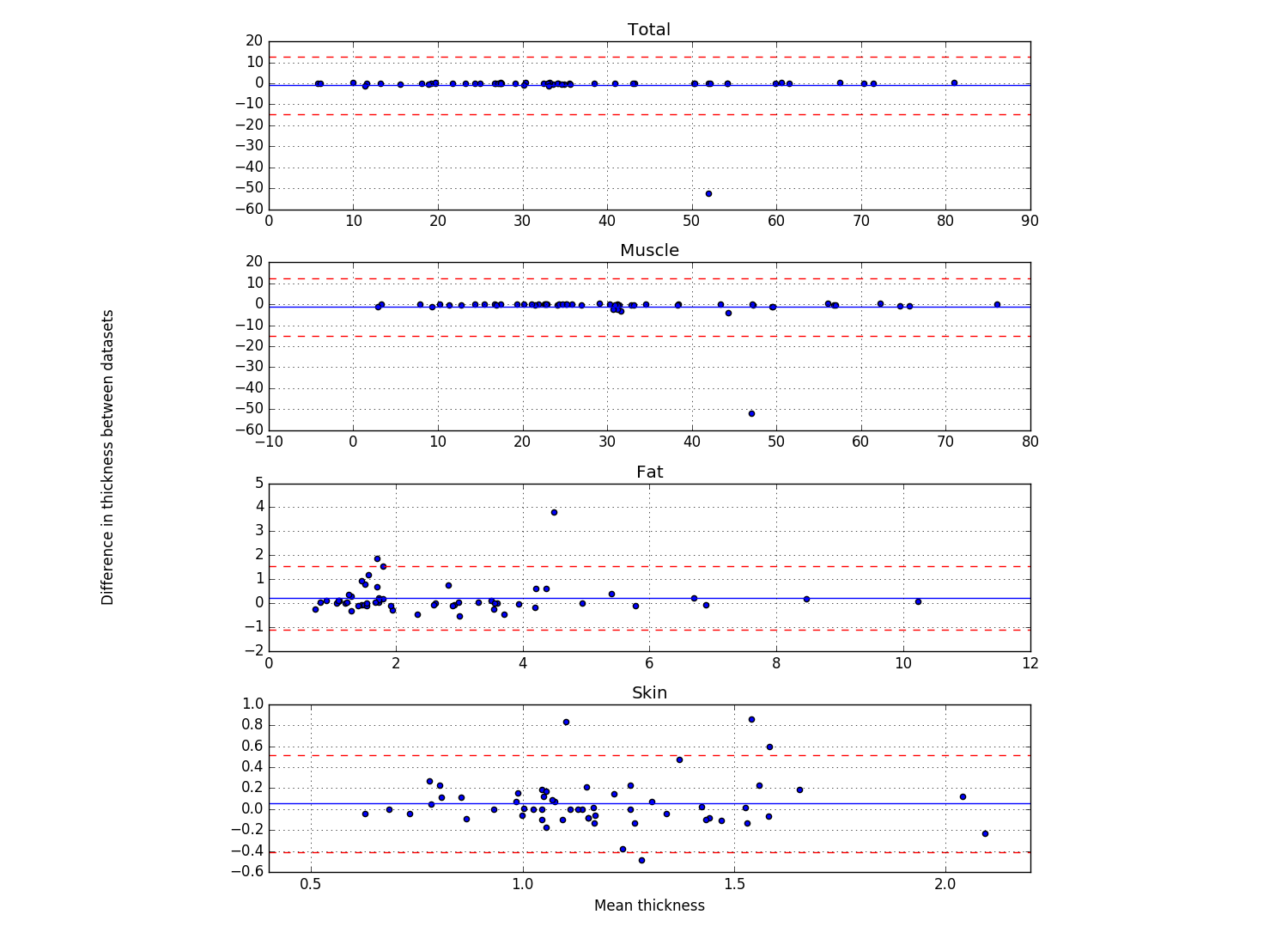
Bland & Altman plots (example below) showing the thicknesses of the first frame of each location that was analyzed (Plot_thicknessForce_reliability_ALL.py)
- Blue line shows the mean difference
- Red lines show 2 standard deviations above and below the mean difference
- Script outputs the trial number of the differences that are further than 2 standard deviations from the mean difference.
Results for reproducibility of MULTIS004 (Rici and Ahmet both analyzed all anatomical trials). Bland Altman Plot
Automated Thickness Measurement Development
- Note: The automated development has been suspended until data is collected on additional subjects.
- Texture analysis of boundaries (transducer/skin, skin/fat, fat/muscle, and muscle/bone)
- Extract rectangles (3mm x 6mm) with boundary located in center
- Analyze by regions
- Fit to a Gaussian curve (average horizontally and then normalize by peak intensity)
- Examine moments of the image using skimage.measure module (Hu's moments, first invariant is analogous to moment of inertia)
Initial results report (using 12 locations from MULTIS-001-1): Specifications/DataAnalysis/report1.pdf
MRI/CT Thickness Analysis
Thickness Analysis Software
Program follows the Ultrasound thickness analysis with several modifications.
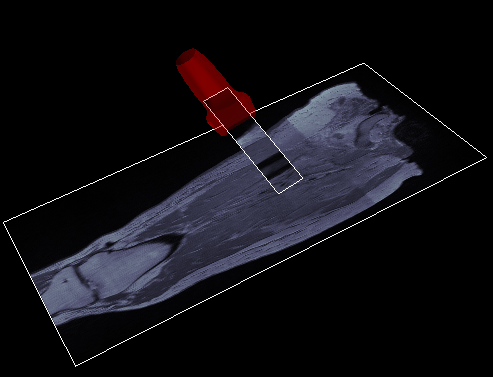
Complete the registration and transformations of ultrasound positions to desired imaging modality coordinate system (MRI or CT). See Specifications/DataAnalysis/Registration page for protocols.
- Necessary files for registration and thickness analysis
- Imaging folder (MRI or CT)
- Ultrasound position xml in desired image coordinate system (MRI or CT)
- CT or MRI image in NIFTI format
- Imaging folder (MRI or CT)
- Required Python scripts
TA_sfm_CT_zProbe.py
TA_sfm_MR_zProbe.py
slice_view.py
tdsmParserMultis.py
XMLparser.py
This script will create 2D slices to represent the ultrasound field of view for CT and MRI
- Install the required libraries as needed
- Change the directory, subject name, modality, and segment at the bottom of the script
Directory (dir) = directory of all in vitro subjects
- Subject (subj_path) = name of subject folder (i.e. CMULTIS002-2)
- Imaging modality (modality) = either CT or MRI
Extremity segment (segment) = either 'UpperLeg', 'LowerLeg', 'UpperArm', or 'LowerArm'
- Run the script.
- Will create .vtk images in a new folder within the subject root folder for each location (CT_Slices or MR_Slices)

Running the manual thickness measurement script. CT script and MRI script
- Install libraries as needed
- Change the directory to match where the Python scripts are located
- Change the filename in getFiles() to the position xml (i.e. CMULTIS002-2_UL_US_CT.xml) created in the registration/transformation step
- Run script
- A list will appear in which will show all the positions for that segment
- Double-click on the location you would like to analyze
- Drag the red dots so that the center of the dot is over each boundary (use the built in matplotlib toolbar to zoom in if needed)
- Transducer/Skin boundary
- Skin/Fat boundary
- Fat/Muscle boundary
- Muscle/Bone boundary
- When you are happy with your selection, press 'Done'. Continue until all locations have been analyzed.
- Data storage
- The xml file will be named using the ultrasound tdms filename with '_manThick'+ time stamp appended
- Version of software used for analysis (TA_sfm.py)
- File Info includes the time stamp at the beginning of analysis as well as the user name at the time of program execution
- Subject shows the MULTIS ID name
- Source shows the Filename of the data (tdms) file
- A new 'Frame' child is appended
- The Frame child includes the following information:
- Value - Image frame/slice number
- Thicknesses - Skin, Fat, and Muscle layers (in mm)
- USPosition - Ultrasound tip position where the thicknesses were analyzed at (coordinate system is recorded)
- The Frame child includes the following information:
- The xml file will be named using the ultrasound tdms filename with '_manThick'+ time stamp appended
Preliminary Results/Questions
In Vivo Thickness Analysis
Report for radiologist examination. Thickness Analysis Questions for Radiologist
- Meeting notes with Dr. Delzell (2016-11-21)
- Images with unsure bone boundaries were difficult to determine if it was bone or fascia, Dr. Delzell suggests using the transverse imaging mode to find the bone first and then switching to the longitudinal mode to capture our images.
- When looking for the boundary of fat to muscle, the fascia of the muscle should be clearly defined by a bright signal (if in the correct plane)
- Many of the distal locations have little to no fat
See Thickness Analysis Comments from Dr. Delzell for more comments relating to specific images.
- Meeting notes with Dr. Delzell (2016-11-21)
In Vitro Ultrasound/MRI Thickness Comparison
Trial 1
- MRI: Trial 4, Ultrasound: MULTIS_C_008-1
- Used ITK snap to estimate MRI thicknesses
- There are quite a few discrepancies between the two imaging modalities
- Initial thoughts:
- Consistency in position of leg between Ultrasound and MRI - Potentially a stand for the head of the femur
- Positions (proximal, central, distal, medial, lateral, anterior, posterior) from the MRI may not match the positions of the Ultrasound - this will be improve once Optotrak is implemented
- Concern with consistency between in vivo and in vitro experimentation - Tyler will work with Tammy to learn the in vivo protocol and apply it to in vitro
- Improvements needed:
- Consistency between ultrasound of in vivo and in vitro
- Consistency between ultrasound and MRI of the in vivo extremities
Trial 2
CMULTIS002-2 (Leg) & CMULTIS002-3 (Arm)
- Leg was taken off of fixture between MRI/CT and Ultrasound imaging
- Arm was imaged with Ultrasound before MRI/CT (irregular workflow)
- Data collection errors resulted in having to re-do the posterior positions during ultrasound data collection, so errors in match-up of imaging modalities are still expected to be high.