Logistics
Schedule |
|||
|
Day 1 |
Day 2 |
Day 3 |
Morning |
Thaw Specimen |
MRI Prep |
Ultrasound Prep |
Mid |
Thaw Specimen |
MRI |
Ultrasound/Anthropometric |
Afternoon |
Thaw Specimen |
CT |
re-freeze |
|
|
refrigerate |
|
* Note: CMULTIS011-1 US/CT/MRI done all on Day 2 |
|||
Target Outcome
- Demographic documentation (age, gender, race, ethnicity)
- Anthropometric measurements (body mass, height, extremity lengths and circumferences)
- A set of MR and CT images of upper and lower leg and arm.
- Ultrasound images at unloaded state to extract tissue thickness for various extremity locations
- Ultrasound imaging during indentation to extract tissue thickness for various extremity locations
- Loads applied by the probe on the segment during unloaded state and indentation
Prerequisites
Specifications
Infrastructure
Protocols
Input
Cadaver upper and lower leg and arm of the same donor
Preparation
Supplies/Equipment needed for experimentation
- Supplies: scalpel, forceps, registration markers (12 for leg or 7 for arm), 2 Optotrak triax clusters, Instrumented Ultrasound, 2 drills, drill bit + tap 8-32 (for registration markers), drill bit (pre-drill for screws), drill bit + tap (for Optotrak base plugs), ultrasound gel, pens, washable markers, cloth tape measure, wash cloths
- Equipment: Optotrak camera system, Optotrak probe, Ultrasound system with foot-switch, 14L5 probe, 9L4 probe, probe casings, load-transducer computer, load-transducer unit, ATI IFPS box, blue DAQ unit, beige DAQ unit, cadaver fixture
Specimen Preparation Imaging and Indentation
One specimen (e.g. Upper and Lower Leg) will be used at a time
Preparing Ultrasound
Refer to in vivo protocol
Deviations from in vivo protocol
- Select 'Most Recent' when prompted to select state file
- Clear state and sensor files to load appropriate appendage state file
load MostRecentState_leg or MostRecentState_arm
load MostRecentSensor_leg or MostRecentSensor_arm
- If trouble connecting Optotrak, check IP address using ipconfig command in CMD Prompt.
- Update the IP address in the configuration file in the Functions folder of the SCCAD tool kit for that appropriate LabVIEW year version
- If vectorNAV times out, unplug and re-plug. It may be necessary to check the COM port using vectorNAV software. Then, update the port in the sensor configuration of LabVIEW.
- It is essential to run a python script after the first few trials to ensure proper connection and transformation. It must also be run after all trials to check optotrak visibility before refreezing the specimen.
in_vitro_experimentCheck_boneCS.py in the Registration folder of the repository
Procedures
Overview
Thaw specimen per guidelines found here
- Tissue Removal
- Remove tissue surrounding femoral head or humeral head to attach specimen to the imaging fixture and insert registration markers.
- Note for the arm, the shoulder had to be dislocated and the arm separated.
- Partially secure specimen to imaging fixture in proper anatomical position, screwing humeral or femoral heads to the custom socket.
- This must happen before registration marker insertion so they do not interfere with the imaging fixture.
- Attach registration markers. 12 (leg), 7 (arm)
- Use the tap with the most available threads to avoid re threading when reversing
- Ensure snug 'wiggle-free' fit of as many markers as possible
- Completely secure specimen to imaging fixture.
- Remove any metal for MRI and CT imaging.
- Transport to MRI, then CT
- Double bag the specimen, now attached to the imaging fixture, using sterile technique
- Refrigerate
- Prep for Ultrasound
- Attach Optotrak triaxial clusters as per the Supplemental Details below.
- Use #4-1 zinc plated wood screws to attach opto base plugs
- Use smallest available drill bit for head regions due to softer bone otherwise screw will spin freely
- Placement must be optimized to maintain constant view with cameras during procedures.
- Connect and configure all sensors and tools
- Optotrak
- Connect all sensors in the following order (MUST be done in order)
- Femur triax (whatever it may be named)
- Tibia triax (whatever it may be named)
- Ultrasound
- Digitizing Probe
- Select 'No' when asked to stream wirelessly
- Establish connection and test visibility of all rigid-bodies
- Digitize the following
- all registration markers
- instrumented ultrasound probe
- imaging fixture
- bone fiducial markers
- Connect all sensors in the following order (MUST be done in order)
- Instrumented Ultrasound
- Perform Weight compensation
- Optotrak
Begin recording digital video with 2 of the GoPro camera
Complete anatomical and indentation trials using the same protocol found in the in vivo experimentation
- Anthropometric measurements
Found here.
- This is done after the specimen is secured in the imaging fixture to create consistency between MRI/CT and manual anthropometric measurements.
- Prepare specimen for refreezing
- Wet with saline, wrap in chucks, double bag
- Refreeze specimen
- At a later date, tissue will be dissected for mechanical testing
Data Transfer
- On the load cell computer, zip the raw data for this subject. The folder can be found in c:\cMULTIS data.
The zipped folder can be transferred to MIDAS (http://cobicore.lerner.ccf.org/midas/community/7) using the secure wireless connection. The data will be saved in the path /Private. The folder may be unzipped, but leave original zipped folder.
- This data will be transferred to the 'MULTIS_trials' folder on the linux computer located in the lab for file association.
- On the ultrasound computer, transfer the ultrasound data in DICOM Format by inserting a USB stick into the ultrasound machine. This USB stick should have a directory named "EXPORT" in the root of the USB drive.
- Press Patient Browser (F2) and select the exam to be transferred.
- Click on the Transfer drop-down menu and select Export to offline.
- Select Drive: E:\Export and Format: DICOM
When the transfer is complete, insert the USB stick into the load cell computer and transfer to the desktop. Zip the ultrasound data and upload the zipped folder to the Ultrasound directory for this subject (http://cobicore.lerner.ccf.org/midas/community/7 and saved in the path /Private).
- This data will also be transferred to the 'MULTIS_trials' folder on the linux computer located in the lab for file association.
- After 2 weeks, the data can be removed from the ultrasound system.
Output
All raw data can be found at http://cobicore.lerner.ccf.org/midas/community/7
- Ultrasound images in DICOM format are located in the path /Private/CMULTISXXX-1/Ultrasound, where XXX is the unique subject identifier (i.e., 001, 002, 003, etc.)
- Configuration and data directories are located in the path /Private/CMULTISXXX-1/, where XXX is the unique subject identifier (i.e., 001, 002, 003, etc.)
Next step: DataAnalysis
Supplemental Details
Optotrak Marker Assembly on Bones
- Mounting of Optotrak markers:
Leg
- Place the femur Optotrak marker on femur base plug
- Place the tibia Optotrak marker on tibia base plug
Arm
- Place the radius Optotrak marker on radius base plug
- Place the humerus Optotrak marker on humerus base plug
Acquisition of Registration Marker Locations and Anatomical Landmarks
Registration marker data collection
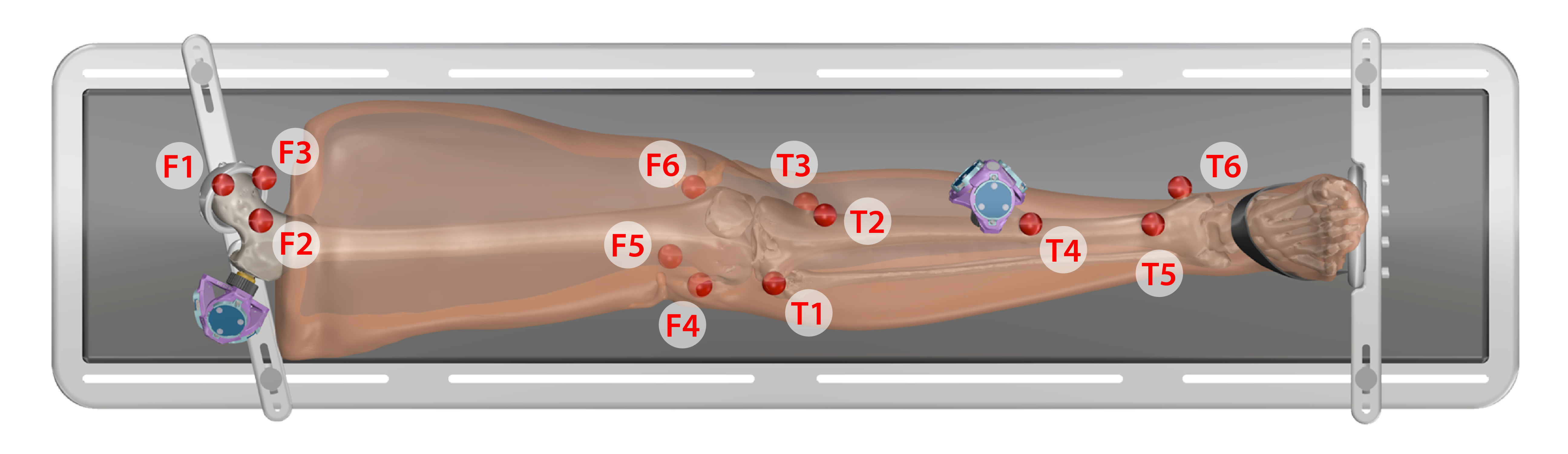
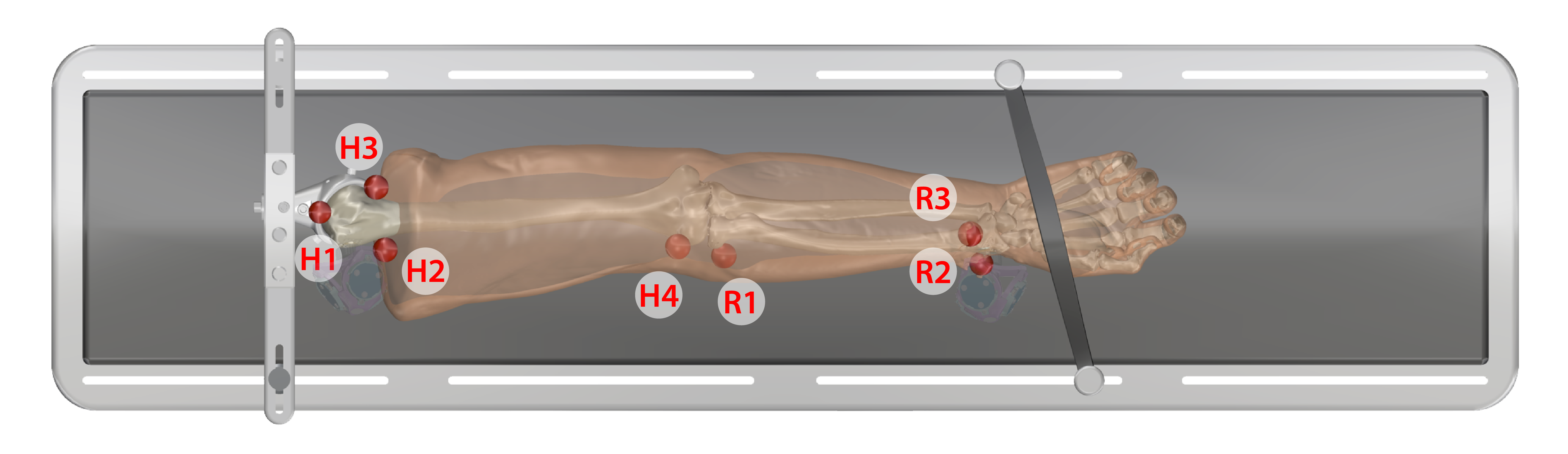
- Keep specimen within Optotrak measurement view.
- Use the digitizing probe to record registration marker locations along with Optotrak marker position/orientation output, measured with respect to the global Optotrak coordinate system for each respective bone
- Ten points on each spherical marker should be digitized such that they are distributed evenly about the sphere surface
- The spheres should be digitized in the following order:
Leg
- Proximal Femur
- F1. Central
- F2. Lateral
- F3. Medial
- Distal Femur
- F4. Lateral epicondyle
- F5. Anterior
- F6. Medial epicondyle
- Proximal Tibia
- T1. Laterial tibial condyle
- T2. Tibial tuberosity
- T3. Medial tibial condyle
- Distal Tibia
- T4. Shaft
- T5. Distal anterior lateral surface
- T6. Medial malleolus
Arm
- Humerus
- H1. Anterior proximal head
- H2. Lateral proximal
- H3. Medial proximal
- H4. Later epicondyle
- Radius
- R1. Neck of radius
- R2. Lateral distal
- R3. Anterior lateral
Anatomical Landmark data collection
- Keep specimen within Optotrak measurement view.
- Use the digitizing probe to record anatomical landmark locations along with Optotrak marker position/orientation output, measured with respect to the global Optotrak coordinate system for each respective bone.
- The following anatomical landmarks will be collected:
Leg
- Femur
- Lateral Femoral Epicondyle
- Medial Femoral Epicondyle
- Femoral Head Point 1
- Femoral Head Point 2
- Femoral Head Point 3
- Femoral Head Point 4
- Tibia
- Lateral Tibial Plateau
- Medial Tibial Plateau
- Lateral Malleolus
- Lateral Malleolus (again)
- Medial Malleolus
- Medial Malleolus (again
Arm
- Humerus
- Lateral Epicondyle
- Medial Epicondyle
- Humeral Head Point
- Ulna
- Lateral Epicondyle
- Medial Epicondyle
- Ulnar Styloid
Note that the specifications dictate that all specimens should be from the right side. If the data collection software is used to collect data from a left specimen, the data file labels will be incorrect as right handed coordinate systems will be created for the left knee. The following chart describes the anatomical meaning for each segment.
Segment Coordinate system definition (Right and Left) |
|||||
Upper/Lower Leg |
Upper/Lower Arm |
||||
|
Left |
Right |
|
Left |
Right |
X |
Medial |
Medial |
X |
Posterior |
Anterior |
Y |
Anterior |
Posterior |
Y |
Superior |
Superior |
Z |
Superior |
Superior |
Y |
Lateral |
Lateral |
Ultrasound Indentation
Refer to in vivo wiki as it is the same protocol.
Optotrak Synchronization
A small time delay exists between Optotrak and force/orientation acquisition. An experiment was performed to quantify the delay for further synchronization.
Experiment
- While the instrumented ultrasound was in view of optotrak, the probe was tapped with enough force to move the device so the movement and force could be viewed simultaneously.
- Tap 3 times
Results
- avg ~43ms +/- 16